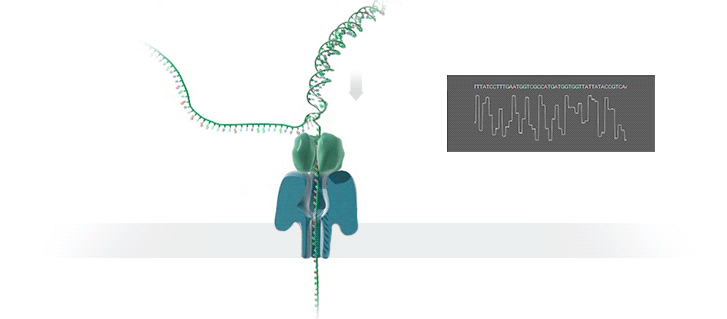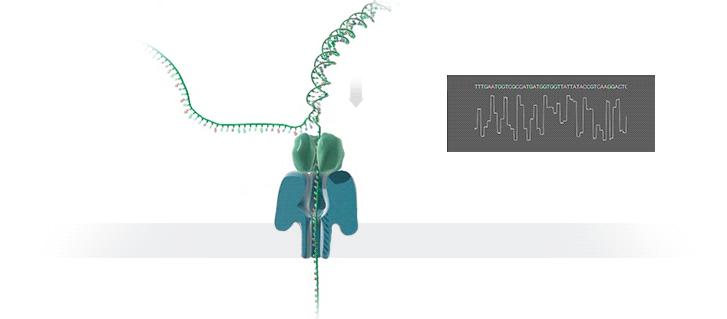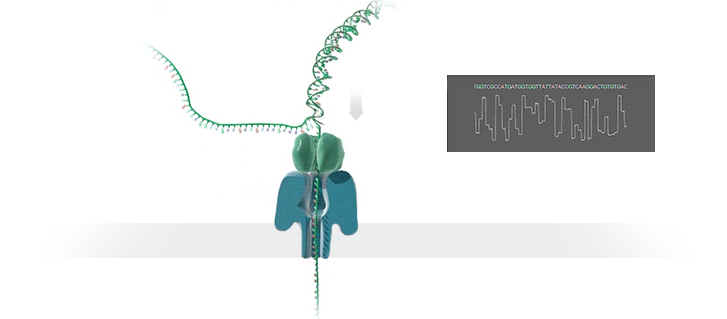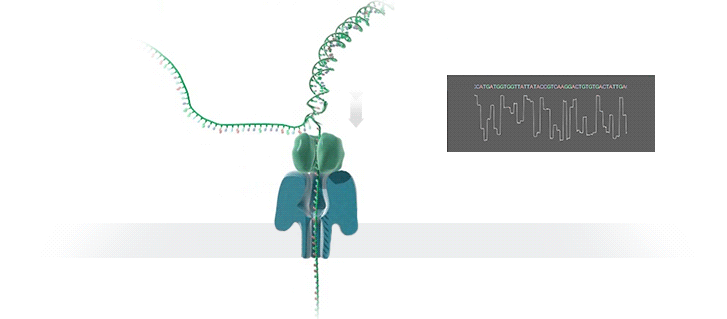
The Human Genome Project took 13 years to complete at a cost of hundreds of millions of dollars. The device described below is available now, costs US$1,000.00 and can sequence an individual’s genome in roughly 3 days. Technological progress is so cool! What a time to be alive!
https://nanoporetech.com/products/minion
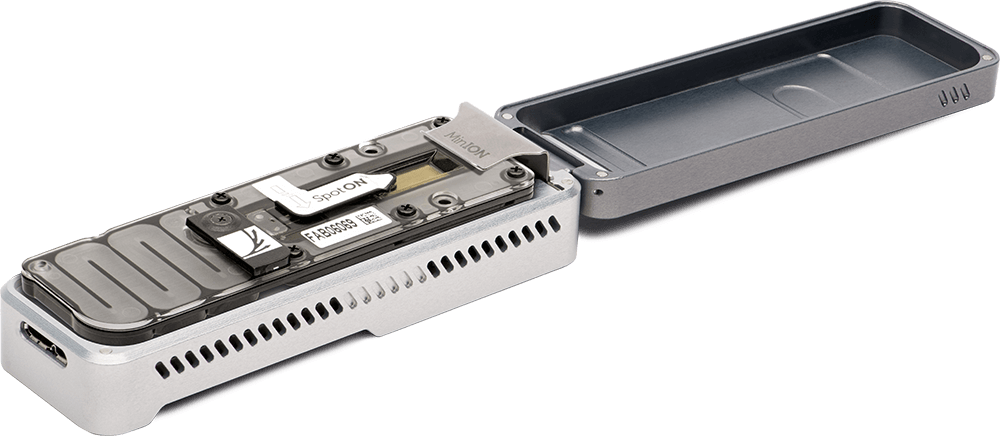
MinION is the only portable real-time device for DNA and RNA sequencing.
Each consumable flow cell can now generate 10–20 Gb of DNA sequence data. Ultra-long read lengths are possible (hundreds of kb) as you can choose your fragment length. The MinION streams data in real time so that analysis can be performed during the experiment and workflows are fully versatile.
The MinION weighs under 100 g and plugs into a PC or laptop using a high-speed USB 3.0 cable. No additional computing infrastructure is required. Not constrained to a laboratory environment, it has been used up a mountain, in a jungle, in the arctic and on the International Space Station.
The MinION is commercially available, simply by paying a starter-pack fee of $1,000. The MinION starter pack includes materials you need to run initial sequencing experiments, including a MinION device, flow cells and kits, as well as membership of the Nanopore Community.
https://www.nature.com/articles/nbt.4060
Discussion
We report sequencing and assembly of a human genome with 99.88% accuracy and an NG50 of 6.4 Mb using unamplified DNA and nanopore reads followed by short-read consensus improvement. At 30× coverage we have produced the most contiguous assembly of a human genome to date, using only a single sequencing technology and the Canu assembler23. Consistent with the view that the underlying ionic raw current contains additional information, signal-based polishing14 improved the assembly accuracy to 99.44%. Finally, we report that combining signal-based polishing and short-read (Illumina) correction26 gave an assembly accuracy of 99.96%, which is similar to metrics for other mammalian genomes9.